Note
Click here to download the full example code
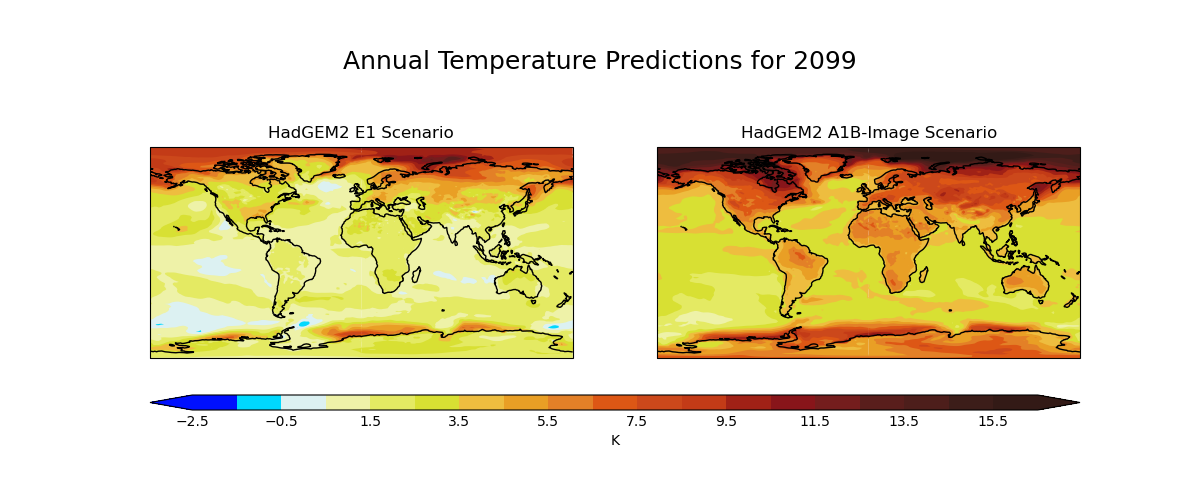
Global Average Annual Temperature Maps
Produces maps of global temperature forecasts from the A1B and E1 scenarios.
The data used comes from the HadGEM2-AO model simulations for the A1B and E1 scenarios, both of which were derived using the IMAGE Integrated Assessment Model (Johns et al. 2011; Lowe et al. 2009).
References
Johns T.C., et al. (2011) Climate change under aggressive mitigation: the ENSEMBLES multi-model experiment. Climate Dynamics, Vol 37, No. 9-10, doi:10.1007/s00382-011-1005-5.
Lowe J.A., C.D. Hewitt, D.P. Van Vuuren, T.C. Johns, E. Stehfest, J-F. Royer, and P. van der Linden, 2009. New Study For Climate Modeling, Analyses, and Scenarios. Eos Trans. AGU, Vol 90, No. 21, doi:10.1029/2009EO210001.
import os.path
import matplotlib.pyplot as plt
import numpy as np
import iris
import iris.coords as coords
import iris.plot as iplt
def cop_metadata_callback(cube, field, filename):
"""
A function which adds an "Experiment" coordinate which comes from the
filename.
"""
# Extract the experiment name (such as A1B or E1) from the filename (in
# this case it is just the start of the file name, before the first ".").
fname = os.path.basename(filename) # filename without path.
experiment_label = fname.split(".")[0]
# Create a coordinate with the experiment label in it...
exp_coord = coords.AuxCoord(
experiment_label, long_name="Experiment", units="no_unit"
)
# ...and add it to the cube.
cube.add_aux_coord(exp_coord)
def main():
# Load E1 and A1B scenarios using the callback to update the metadata.
scenario_files = [
iris.sample_data_path(fname) for fname in ["E1.2098.pp", "A1B.2098.pp"]
]
scenarios = iris.load(scenario_files, callback=cop_metadata_callback)
# Load the preindustrial reference data.
preindustrial = iris.load_cube(iris.sample_data_path("pre-industrial.pp"))
# Define evenly spaced contour levels: -2.5, -1.5, ... 15.5, 16.5 with the
# specific colours.
levels = np.arange(20) - 2.5
red = (
np.array(
[
0,
0,
221,
239,
229,
217,
239,
234,
228,
222,
205,
196,
161,
137,
116,
89,
77,
60,
51,
]
)
/ 256.0
)
green = (
np.array(
[
16,
217,
242,
243,
235,
225,
190,
160,
128,
87,
72,
59,
33,
21,
29,
30,
30,
29,
26,
]
)
/ 256.0
)
blue = (
np.array(
[
255,
255,
243,
169,
99,
51,
63,
37,
39,
21,
27,
23,
22,
26,
29,
28,
27,
25,
22,
]
)
/ 256.0
)
# Put those colours into an array which can be passed to contourf as the
# specific colours for each level.
colors = np.stack([red, green, blue], axis=1)
# Make a wider than normal figure to house two maps side-by-side.
fig, ax_array = plt.subplots(1, 2, figsize=(12, 5))
# Loop over our scenarios to make a plot for each.
for ax, experiment, label in zip(
ax_array, ["E1", "A1B"], ["E1", "A1B-Image"]
):
exp_cube = scenarios.extract_cube(
iris.Constraint(Experiment=experiment)
)
time_coord = exp_cube.coord("time")
# Calculate the difference from the preindustial control run.
exp_anom_cube = exp_cube - preindustrial
# Plot this anomaly.
plt.sca(ax)
ax.set_title(f"HadGEM2 {label} Scenario", fontsize=10)
contour_result = iplt.contourf(
exp_anom_cube, levels, colors=colors, extend="both"
)
plt.gca().coastlines()
# Now add a colourbar who's leftmost point is the same as the leftmost
# point of the left hand plot and rightmost point is the rightmost
# point of the right hand plot.
# Get the positions of the 2nd plot and the left position of the 1st plot.
left, bottom, width, height = ax_array[1].get_position().bounds
first_plot_left = ax_array[0].get_position().bounds[0]
# The width of the colorbar should now be simple.
width = left - first_plot_left + width
# Add axes to the figure, to place the colour bar.
colorbar_axes = fig.add_axes([first_plot_left, 0.18, width, 0.03])
# Add the colour bar.
cbar = plt.colorbar(
contour_result, colorbar_axes, orientation="horizontal"
)
# Label the colour bar and add ticks.
cbar.set_label(preindustrial.units)
cbar.ax.tick_params(length=0)
# Get the time datetime from the coordinate.
time = time_coord.units.num2date(time_coord.points[0])
# Set a title for the entire figure, using the year from the datetime
# object. Also, set the y value for the title so that it is not tight to
# the top of the plot.
fig.suptitle(
f"Annual Temperature Predictions for {time.year}",
y=0.9,
fontsize=18,
)
iplt.show()
if __name__ == "__main__":
main()
Total running time of the script: ( 0 minutes 2.019 seconds)