3. Dask Bags and Greedy Parallelism#
Here is a journey that demonstrates:
How to apply dask.bags to an existing script
The equal importance of optimisation of non-parallel parts of a script
Protection against multiple softwares trying to manage parallelism simultaneously
3.1 The Problem - Slow Loading#
We have ~7000 GRIB files spread between 256 dated directories:
.
|-- 20180401
| |-- gfs.t00z.icing.0p25.grb2f006
| |-- gfs.t00z.icing.0p25.grb2f006.1
| |-- gfs.t00z.icing.0p25.grb2f012
| |-- gfs.t00z.icing.0p25.grb2f018
| |-- gfs.t00z.icing.0p25.grb2f024
| |-- gfs.t00z.icing.0p25.grb2f030
| `-- gfs.t00z.icing.0p25.grb2f036
|-- 20180402
| `-- gfs.t00z.icing.0p25.grb2f006
|-- 20180403
| |-- gfs.t12z.icing.0p25.grb2f006
| |-- gfs.t12z.icing.0p25.grb2f012
With this script, a sample of 11 GRIB files takes ~600secs to load:
import iris
import glob
fpaths=glob.glob('20190416/*t18z*f???')
cubes = iris.load(fpaths, callback=callback)
def callback(cube, field, fname):
if field.sections[5]['bitsPerValue'] == 0:
raise iris.exceptions.IgnoreCubeException
if field.sections[4]['parameterNumber'] == 20:
raise iris.exceptions.IgnoreCubeException
elif field.sections[4]['parameterNumber'] == 234:
cube.long_name = 'Icing Severity'
3.2 Parallelisation#
We’ll try using dask.bag to parallelise the function calls. It’s important that Dask is given the freedom to break the task down in an efficient manner - the function that is mapped across the bag should only load a single file, and the bag itself can iterate through the list of files. Here’s the restructured script:
import glob
import multiprocessing
import os
import dask
import dask.bag as db
import iris
def callback(cube, field, fname):
if field.sections[5]['bitsPerValue'] == 0:
raise iris.exceptions.IgnoreCubeException
if field.sections[4]['parameterNumber'] == 20:
raise iris.exceptions.IgnoreCubeException
elif field.sections[4]['parameterNumber'] == 234:
cube.long_name = 'Icing Severity'
def func(fname):
return iris.load_cube(fname, callback=callback)
fpaths = list(glob.glob('20190416/*t18z*f???'))
# Determine the number of processors visible ..
cpu_count = multiprocessing.cpu_count()
# .. or as given by slurm allocation.
# Only relevant when using Slurm for job scheduling
if 'SLURM_NTASKS' in os.environ:
cpu_count = os.environ['SLURM_NTASKS']
# Do not exceed the number of CPUs available, leaving 1 for the system.
num_workers = cpu_count - 1
print('Using {} workers from {} CPUs...'.format(num_workers, cpu_count))
# Now do the parallel load.
with dask.config.set(num_workers=num_workers):
bag = db.from_sequence(fpaths).map(func)
cubes = iris.cube.CubeList(bag.compute()).merge()
This achieves approximately a 10-fold improvement if enough CPUs are available to have one per file. See this benchmarking:
Machine |
CPUs Available |
CPUs Used |
Time Taken |
|---|---|---|---|
A |
4 |
3 |
4m 05s |
4 |
3m 22s |
||
B |
8 |
1 |
9m 10s |
7 |
2m 35s |
||
8 |
2m 20s |
3.3 Profiling#
1m 10s is still a surprisingly long time. When faced with a mystery like this it is helpful to profile the script to see if there are any steps that are taking more time than we would expect. For this we use a tool called kapture to produce a flame chart visualising the time spent performing each call:
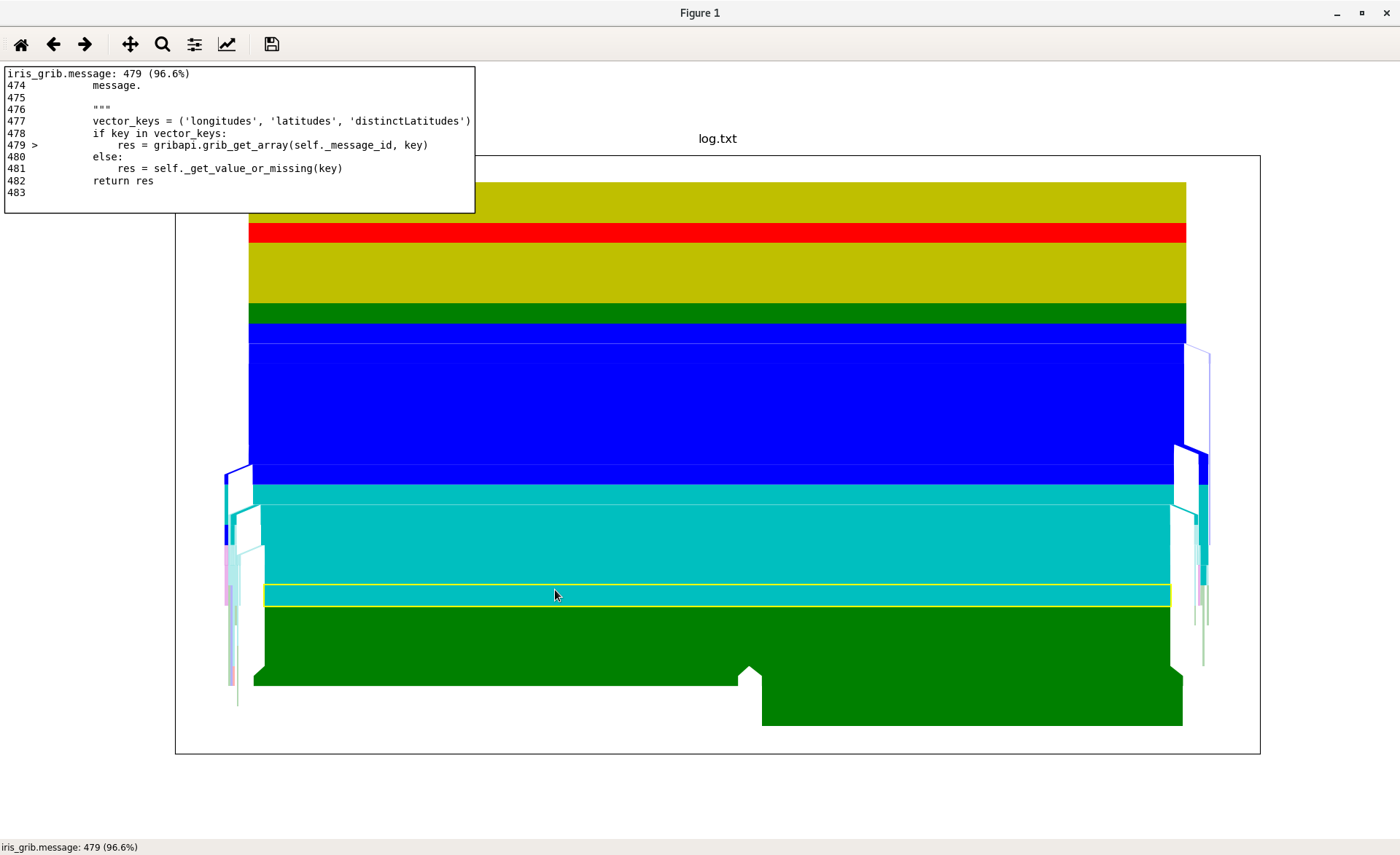
From this we see that 96% of the runtime is taken by this call:
res = gribapi.grib_get_array(self._message_id, key)
This is the call being used during the callback function when it uses
GRIB messages to filter out cubes with certain unwanted properties.
3.4 Improving GRIB Key Handling#
Even with parallelisation, we are still limited by the time it takes to run a single instance of a function. This is going to become much more important when running 7000 files instead of 11, since there will be nowhere near enough CPUs even on a large multi-processing system, meaning each CPU will be running many instances of the function. Parallelisation can only go so far to solving speed issues – it’s effectively the ‘brute force’ method.
3.3 Profiling showed us where the major bottleneck is. To improve efficiency we can re-write the script to filter on GRIB messages before converting the GRIB file to a cube:
import dask
import dask.bag as db
import glob
import iris
import multiprocessing
import os
def func(fname):
import iris
from iris_grib import load_pairs_from_fields
from iris_grib.message import GribMessage # perform GRIB message level filtering...
filtered_messages = []
for message in GribMessage.messages_from_filename(fname):
if (message.sections[5]['bitsPerValue'] != 0 and
message.sections[4]['parameterNumber'] == 234):
filtered_messages.append(message) # now convert the messages to cubes...
cubes = [cube for cube, message in load_pairs_from_fields(filtered_messages)]
return iris.cube.CubeList(cubes).merge_cube()
fpaths = list(glob.glob('/scratch/frcz/ICING/GFS_DATA/20190416/*t18z*f???'))
cpu_count = multiprocessing.cpu_count()
# Only relevant when using Slurm for job scheduling
if 'SLURM_NTASKS' in os.environ:
cpu_count = os.environ['SLURM_NTASKS']
num_workers = cpu_count - 1
print('Using {} workers from {} CPUs...'.format(num_workers, cpu_count))
with dask.config.set(num_workers=num_workers):
bag = db.from_sequence(fpaths).map(func)
cubes = iris.cube.CubeList(bag.compute())
This achieves a significant performance improvement - more than twice as fast as the previous benchmarks:
Machine |
CPUs Available |
CPUs Used |
Previous Time |
New Time |
|---|---|---|---|---|
Example |
8 |
7 |
2m 35s |
1m 05s |
8 |
2m 20s |
1m 03s |
3.5 Managing External Factors#
The speed will still need to be further improved before we can process 7000 files. The main gains we can achieve are by making sure it is only Dask that manages multi-processing - if multi-processing is coming from more than one place there are predictable clashes.
First, NumPy must be prevented from performing it’s own multi-processing by
adding the following before import numpy is called. You can read more
about this in NumPy Threads.
import os
os.environ["OMP_NUM_THREADS"] = "1"
os.environ["OPENBLAS_NUM_THREADS"] = "1"
os.environ["MKL_NUM_THREADS"] = "1"
os.environ["VECLIB_MAXIMUM_THREADS"] = "1"
os.environ["NUMEXPR_NUM_THREADS"] = "1"
Lastly, if you are using SLURM on the computing cluster then SLURM must be configured to prevent it
optimising the number of cores necessary for the job. See the SLURM commands
below, to be added before running the python script. It’s important that
ntasks matches the number of CPUs specified in the python script. You
can read more about these points in CPU Allocation.
#SBATCH --ntasks=12
#SBATCH --ntasks-per-core=1
This has all been based on a real example. Once all the above had been set up correctly, the completion time had dropped from an estimated 55 days to less than 1 day.
3.6 Lessons#
Dask isn’t a magic switch - it’s important to write your script so that there is a way to create small sub-tasks. In this case by providing dask.bag with the file list and the function separated
Parallelism is not the only performance improvement to try - the script will still be slow if the individual function is slow
All multi-processing needs to be managed by Dask. Several other factors may introduce multi-processing and these need to be configured not to
-3.9.0-green?style=flat)